Gene function
Next generation sequencing (NGS) leads to the discovery of many new genetic variants, as outlined by Volker Lauschke et al. in Requirements for comprehensive pharmacogenetic genotyping platforms.
A challenge is how to evaluate the functional consequences of these mutations. A discussion of this is given by Ingelman-Sundberg et al. in the article Integrating rare genetic variants into pharmacogenetic drug response predictions. Another article by the same authors is Prediction of drug response and adverse drug reactions: From twin studies to Next Generation Sequencing
Description of problem
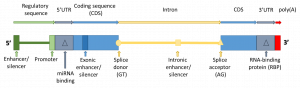
The effects of mutations depend on the region in which they are encountered[1].
Disease causing mutations are often shared by a very small number of people due to evolutionary pressure. For genes involved in drug transport and metabolization, the evolutionary pressure is much weaker, and therefore the variants may be common.
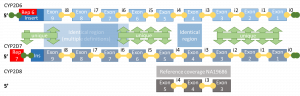
An additional difficulty with determining genetic effect is for larger structural variants caused by copy number variations, pseudogenes or repeated regions. A good example of this is CYP2D6.
Computational methods
A review of computational methods for PGx interpretation of NGS data has been written by Yitian Zhou. Zhou et al. also proposes a tool that combines several such methods, An optimized prediction framework to assess the functional impact of pharmacogenetic variants.
Experimental methods
Ingelman-Sundberg et al. also propose a high-throughput in vitro method for gene function evaluation in Human liver spheroids in chemically defined conditions for studies of gene–drug, drug–drug and disease–drug interactions. This idea is the basis of the company HepaPredict AB.
- ↑ A good description is given by Zouh et al.